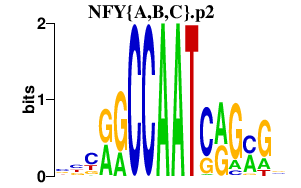
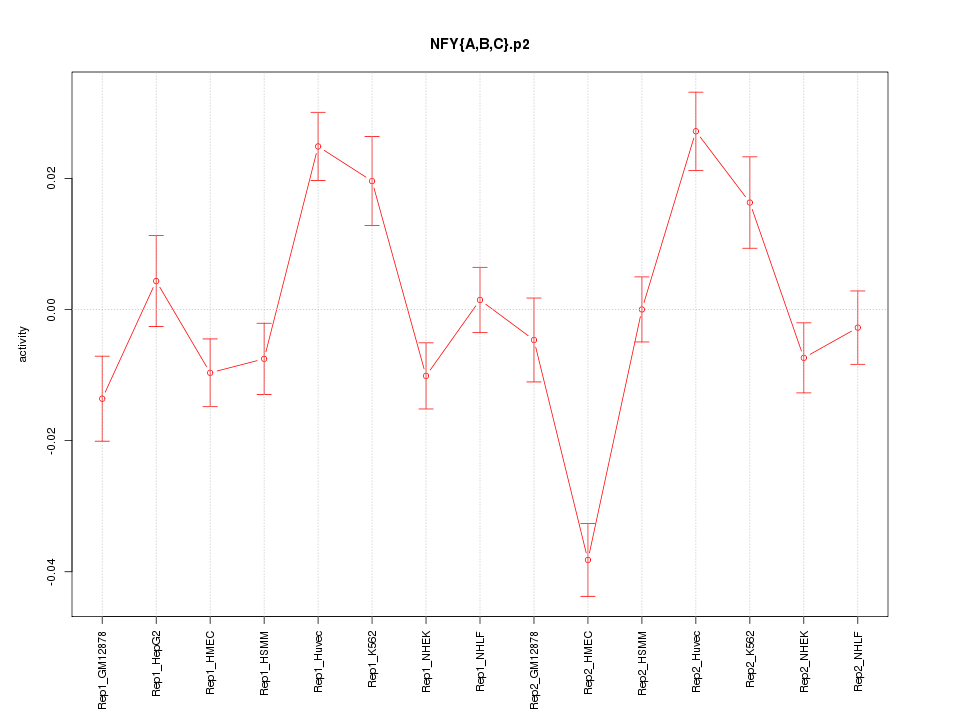
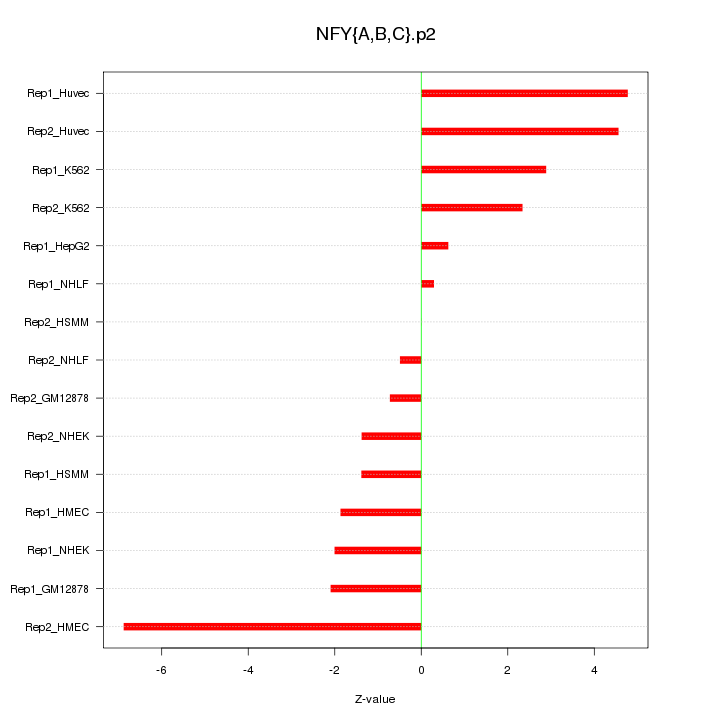
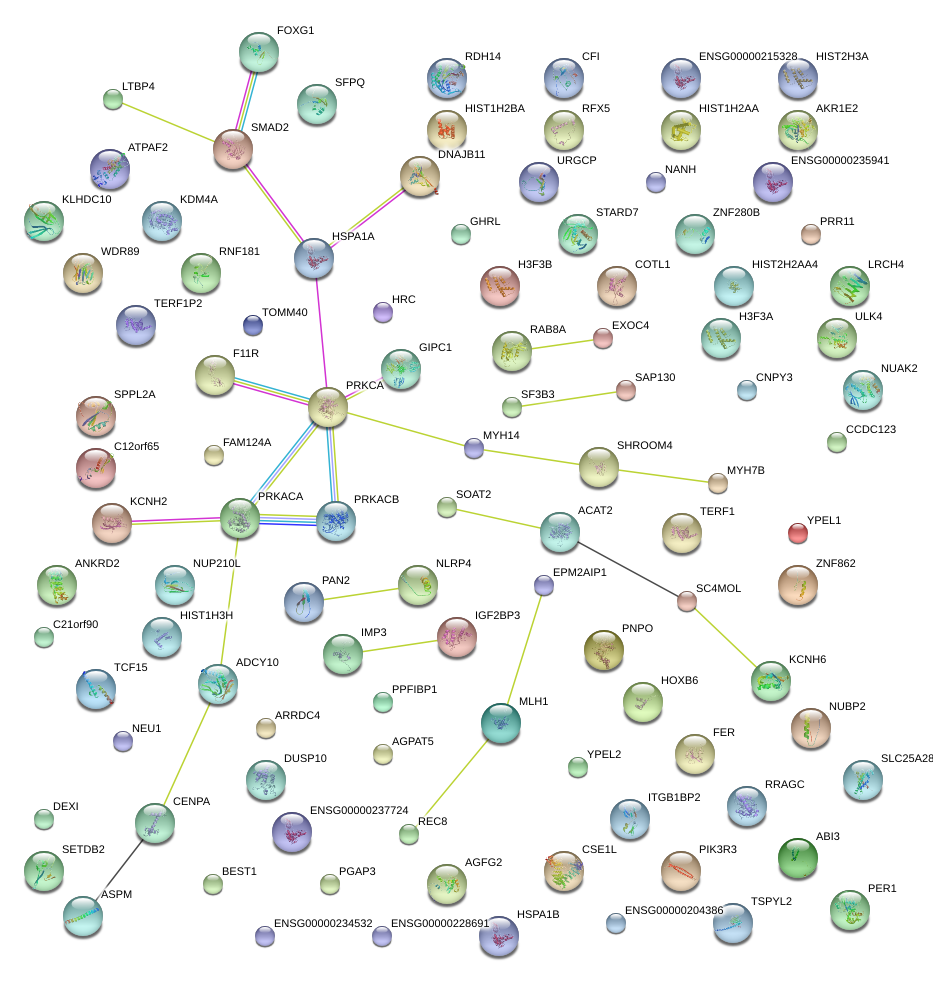
|
chr3_-_41978464
|
1.932
|
NM_017886
|
ULK4
|
unc-51-like kinase 4 (C. elegans)
|
|
chr5_+_108112537
|
1.920
|
|
FER
|
fer (fps/fes related) tyrosine kinase
|
|
chr7_+_129497637
|
1.659
|
|
KLHDC10
|
kelch domain containing 10
|
|
chr19_-_14467939
|
1.650
|
NM_005716
NM_202467
NM_202468
NM_202469
NM_202470
NM_202494
|
GIPC1
|
GIPC PDZ domain containing family, member 1
|
|
chr11_+_61473931
|
1.604
|
NM_001139443
NM_004183
|
BEST1
|
bestrophin 1
|
|
chr19_-_14467890
|
1.597
|
|
GIPC1
|
GIPC PDZ domain containing family, member 1
|
|
chr7_+_129497572
|
1.591
|
NM_014997
|
KLHDC10
|
kelch domain containing 10
|
|
chr15_-_73719546
|
1.571
|
NM_018285
|
IMP3
|
IMP3, U3 small nucleolar ribonucleoprotein, homolog (yeast)
|
|
chr1_-_46370837
|
1.559
|
NM_003629
NM_001114172
|
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma)
|
|
chr1_-_152394215
|
1.506
|
NM_001159484
NM_207308
|
NUP210L
|
nucleoporin 210kDa-like
|
|
chr6_+_160103500
|
1.479
|
|
ACAT2
|
acetyl-CoA acetyltransferase 2
|
|
chr22_-_21193037
|
1.366
|
|
ZNF280B
|
zinc finger protein 280B
|
|
chrX_+_70438352
|
1.320
|
NM_012278
|
ITGB1BP2
|
integrin beta 1 binding protein (melusin) 2
|
|
chr2_+_85676347
|
1.320
|
NM_016494
|
RNF181
|
ring finger protein 181
|
|
chr7_+_129497808
|
1.318
|
|
KLHDC10
|
kelch domain containing 10
|
|
chr1_-_159257486
|
1.317
|
|
F11R
|
F11 receptor
|
|
chr2_-_96237665
|
1.314
|
|
STARD7
|
StAR-related lipid transfer (START) domain containing 7
|
|
chr6_-_25834733
|
1.291
|
NM_170745
|
HIST1H2AA
|
histone cluster 1, H2aa
|
|
chr6_+_25834983
|
1.284
|
NM_170610
|
HIST1H2BA
|
histone cluster 1, H2ba
|
|
chr6_+_31903374
|
1.244
|
NM_005346
|
HSPA1A
HSPA1B
|
heat shock 70kDa protein 1A
heat shock 70kDa protein 1B
|
|
chr21_+_44761927
|
1.229
|
|
C21orf90
|
chromosome 21 open reading frame 90
|
|
chr20_+_47096189
|
1.219
|
NM_001316
|
CSE1L
|
CSE1 chromosome segregation 1-like (yeast)
|
|
chr20_+_47096280
|
1.195
|
|
CSE1L
|
CSE1 chromosome segregation 1-like (yeast)
|
|
chr13_+_48916420
|
1.172
|
NM_031915
|
SETDB2
|
SET domain, bifurcated 2
|
|
chr19_+_45794895
|
1.172
|
NM_001042544
|
LTBP4
|
latent transforming growth factor beta binding protein 4
|
|
chr4_+_166468266
|
1.150
|
NM_001017369
NM_006745
|
SC4MOL
|
sterol-C4-methyl oxidase-like
|
|
chrX_+_53128207
|
1.140
|
NM_022117
|
TSPYL2
|
TSPY-like 2
|
|
chr1_+_43888415
|
1.115
|
|
KDM4A
|
lysine (K)-specific demethylase 4A
|
|
chr2_+_26862416
|
1.107
|
|
CENPA
|
centromere protein A
|
|
chr16_-_10943656
|
1.106
|
|
DEXI
|
Dexi homolog (mouse)
|
|
chr13_+_48916390
|
1.095
|
|
SETDB2
|
SET domain, bifurcated 2
|
|
chr7_-_100021658
|
1.093
|
|
LRCH4
|
leucine-rich repeats and calponin homology (CH) domain containing 4
|
|
chr6_+_31891509
|
1.090
|
|
HSPA1A
HSPA1B
|
heat shock 70kDa protein 1A
heat shock 70kDa protein 1B
|
|
chr1_-_219982016
|
1.072
|
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr22_-_20419975
|
1.064
|
NM_013313
|
YPEL1
|
yippee-like 1 (Drosophila)
|
|
chr8_+_74083615
|
1.058
|
NM_003218
NM_017489
|
TERF1
|
telomeric repeat binding factor (NIMA-interacting) 1
|
|
chr15_+_96304911
|
1.055
|
NM_183376
|
ARRDC4
|
arrestin domain containing 4
|
|
chr1_-_159257595
|
1.053
|
NM_016946
|
F11R
|
F11 receptor
|
|
chr18_-_43711143
|
1.047
|
|
SMAD2
|
SMAD family member 2
|
|
chr1_-_219982049
|
1.035
|
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr1_-_203557379
|
1.034
|
NM_030952
|
NUAK2
|
NUAK family, SNF1-like kinase, 2
|
|
chr17_+_70256518
|
1.033
|
|
|
|
|
chr13_+_48916526
|
1.032
|
|
SETDB2
|
SET domain, bifurcated 2
|
|
chr17_+_54587882
|
0.988
|
|
PRR11
|
proline rich 11
|
|
chr20_-_538909
|
0.988
|
NM_004609
|
TCF15
|
transcription factor 15 (basic helix-loop-helix)
|
|
chr19_+_16083706
|
0.974
|
|
RAB8A
|
RAB8A, member RAS oncogene family
|
|
chr1_-_159257567
|
0.973
|
|
F11R
|
F11 receptor
|
|
chr14_+_23711044
|
0.967
|
NM_001048205
NM_005132
|
REC8
|
REC8 homolog (yeast)
|
|
chr6_+_43005214
|
0.964
|
|
CNPY3
|
canopy 3 homolog (zebrafish)
|
|
chr17_-_17883051
|
0.944
|
NM_145691
|
ATPAF2
|
ATP synthase mitochondrial F1 complex assembly factor 2
|
|
chr7_-_150305933
|
0.942
|
NM_000238
NM_172056
|
KCNH2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2
|
|
chr17_-_8000362
|
0.942
|
|
PER1
|
period homolog 1 (Drosophila)
|
|
chr10_+_4858321
|
0.940
|
NM_001040177
|
AKR1E2
|
aldo-keto reductase family 1, member E2
|
|
chr15_+_96304942
|
0.937
|
|
ARRDC4
|
arrestin domain containing 4
|
|
chr1_-_195382189
|
0.924
|
NM_018136
|
ASPM
|
asp (abnormal spindle) homolog, microcephaly associated (Drosophila)
|
|
chr2_-_128501206
|
0.904
|
|
SAP130
|
Sin3A-associated protein, 130kDa
|
|
chr12_+_27568311
|
0.896
|
NM_001198915
NM_001198916
NM_003622
NM_177444
|
PPFIBP1
|
PTPRF interacting protein, binding protein 1 (liprin beta 1)
|
|
chr7_+_99974781
|
0.895
|
|
AGFG2
|
ArfGAP with FG repeats 2
|
|
chr8_+_6553543
|
0.888
|
|
AGPAT5
|
1-acylglycerol-3-phosphate O-acyltransferase 5 (lysophosphatidic acid acyltransferase, epsilon)
|
|
chr1_-_39097904
|
0.881
|
NM_022157
|
RRAGC
|
Ras-related GTP binding C
|
|
chr10_-_101370076
|
0.880
|
|
SLC25A28
|
solute carrier family 25, member 28
|
|
chr1_-_219982081
|
0.879
|
NM_007207
NM_144729
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr3_-_37009571
|
0.879
|
|
|
|
|
chr16_-_10943716
|
0.862
|
NM_014015
|
DEXI
|
Dexi homolog (mouse)
|
|
chr19_-_54350457
|
0.861
|
NM_002152
|
HRC
|
histidine rich calcium binding protein
|
|
chr12_+_122283796
|
0.859
|
NM_152269
NM_001194995
|
C12orf65
|
chromosome 12 open reading frame 65
|
|
chr17_+_44643067
|
0.857
|
|
ABI3
|
ABI family, member 3
|
|
chr6_-_31938530
|
0.846
|
|
NEU1
|
sialidase 1 (lysosomal sialidase)
|
|
chr14_+_28305922
|
0.846
|
NM_005249
|
FOXG1
|
forkhead box G1
|
|
chr1_+_43888391
|
0.842
|
|
KDM4A
|
lysine (K)-specific demethylase 4A
|
|
chr19_+_55398680
|
0.839
|
NM_001077186
NM_001145809
NM_024729
|
MYH14
|
myosin, heavy chain 14, non-muscle
|
|
chr3_+_187771192
|
0.838
|
|
DNAJB11
|
DnaJ (Hsp40) homolog, subfamily B, member 11
|
|
chr19_-_14089362
|
0.826
|
NM_002730
|
PRKACA
|
protein kinase, cAMP-dependent, catalytic, alpha
|
|
chr1_-_35431318
|
0.826
|
|
SFPQ
|
splicing factor proline/glutamine-rich
|
|
chr3_-_37009592
|
0.822
|
|
EPM2AIP1
|
EPM2A (laforin) interacting protein 1
|
|
chr16_-_83209185
|
0.821
|
|
COTL1
|
coactosin-like 1 (Dictyostelium)
|
|
chr4_-_110942783
|
0.815
|
NM_000204
|
CFI
|
complement factor I
|
|
chr14_-_63178287
|
0.809
|
NM_080666
|
WDR89
|
WD repeat domain 89
|
|
chr19_-_38154631
|
0.807
|
NM_032816
|
CCDC123
|
coiled-coil domain containing 123
|
|
chr16_+_1772929
|
0.801
|
NM_012225
|
NUBP2
|
nucleotide binding protein 2 (MinD homolog, E. coli)
|
|
chr1_-_166149867
|
0.798
|
NM_001167749
NM_018417
|
ADCY10
|
adenylate cyclase 10 (soluble)
|
|
chr3_+_37009987
|
0.793
|
|
MLH1
|
mutL homolog 1, colon cancer, nonpolyposis type 2 (E. coli)
|
|
chr17_-_44037332
|
0.793
|
NM_018952
|
HOXB6
|
homeobox B6
|
|
chr7_+_149166411
|
0.793
|
|
ZNF862
|
zinc finger protein 862
|
|
chr1_-_35431329
|
0.792
|
NM_005066
|
SFPQ
|
splicing factor proline/glutamine-rich
|
|
chr17_+_43374021
|
0.790
|
|
PNPO
|
pyridoxamine 5'-phosphate oxidase
|
|
chr1_-_149586298
|
0.787
|
|
RFX5
|
regulatory factor X, 5 (influences HLA class II expression)
|
|
chr12_-_55013987
|
0.783
|
NM_001166279
NM_014871
|
PAN2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae)
|
|
chr6_-_31938572
|
0.780
|
NM_000434
|
NEU1
|
sialidase 1 (lysosomal sialidase)
|
|
chr19_+_50086299
|
0.778
|
NM_001128916
NM_001128917
NM_006114
|
TOMM40
|
translocase of outer mitochondrial membrane 40 homolog (yeast)
|
|
chrX_-_50403412
|
0.777
|
|
SHROOM4
|
shroom family member 4
|
|
chr17_-_71287417
|
0.767
|
NM_005324
|
H3F3B
|
H3 histone, family 3B (H3.3B)
|
|
chr12_+_88627601
|
0.766
|
|
LOC338758
|
hypothetical LOC338758
|
|
chr1_-_28113817
|
0.763
|
NM_002946
|
RPA2
|
replication protein A2, 32kDa
|
|
chr19_-_12858956
|
0.754
|
NM_006563
|
KLF1
|
Kruppel-like factor 1 (erythroid)
|
|
chr1_-_109742020
|
0.741
|
NM_002959
|
SORT1
|
sortilin 1
|
|
chr6_+_15354278
|
0.740
|
NM_004973
|
JARID2
|
jumonji, AT rich interactive domain 2
|
|
chr16_-_83209140
|
0.737
|
NM_021149
|
COTL1
|
coactosin-like 1 (Dictyostelium)
|
|
chr3_+_199171467
|
0.736
|
NM_001136049
NM_033029
|
LMLN
|
leishmanolysin-like (metallopeptidase M8 family)
|
|
chr17_-_77411733
|
0.735
|
|
P4HB
|
prolyl 4-hydroxylase, beta polypeptide
|
|
chr7_+_99974720
|
0.730
|
NM_006076
|
AGFG2
|
ArfGAP with FG repeats 2
|
|
chr2_-_97646758
|
0.728
|
|
ACTR1B
|
ARP1 actin-related protein 1 homolog B, centractin beta (yeast)
|
|
chr8_+_11697929
|
0.727
|
|
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1
|
|
chr16_+_1773004
|
0.725
|
|
NUBP2
|
nucleotide binding protein 2 (MinD homolog, E. coli)
|
|
chrX_+_47999740
|
0.725
|
NM_005635
|
SSX1
|
synovial sarcoma, X breakpoint 1
|
|
chr8_+_6553278
|
0.724
|
NM_018361
|
AGPAT5
|
1-acylglycerol-3-phosphate O-acyltransferase 5 (lysophosphatidic acid acyltransferase, epsilon)
|
|
chr2_-_61618898
|
0.724
|
NM_003400
|
XPO1
|
exportin 1 (CRM1 homolog, yeast)
|
|
chr2_-_128501334
|
0.719
|
NM_001145928
NM_024545
|
SAP130
|
Sin3A-associated protein, 130kDa
|
|
chr6_+_31891293
|
0.719
|
|
HSPA1A
|
heat shock 70kDa protein 1A
|
|
chr3_-_130385454
|
0.716
|
NM_001127192
NM_001127193
NM_001127194
NM_001127195
NM_001127196
NM_003418
|
CNBP
|
CCHC-type zinc finger, nucleic acid binding protein
|
|
chr19_-_6688270
|
0.716
|
NM_020171
|
GPR108
|
G protein-coupled receptor 108
|
|
chr7_-_135063461
|
0.711
|
NM_012450
|
SLC13A4
|
solute carrier family 13 (sodium/sulfate symporters), member 4
|
|
chr22_+_22737753
|
0.710
|
NM_012295
|
CABIN1
|
calcineurin binding protein 1
|
|
chr4_-_110942576
|
0.708
|
|
CFI
|
complement factor I
|
|
chr17_-_77411695
|
0.708
|
|
P4HB
|
prolyl 4-hydroxylase, beta polypeptide
|
|
chr9_+_132990834
|
0.705
|
|
NUP214
|
nucleoporin 214kDa
|
|
chr9_-_127043169
|
0.701
|
|
HSPA5
|
heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa)
|
|
chr16_-_83209020
|
0.700
|
|
COTL1
|
coactosin-like 1 (Dictyostelium)
|
|
chr19_-_14089560
|
0.698
|
|
PRKACA
|
protein kinase, cAMP-dependent, catalytic, alpha
|
|
chr9_-_35804997
|
0.697
|
|
HINT2
|
histidine triad nucleotide binding protein 2
|
|
chr1_-_35431017
|
0.697
|
|
SFPQ
|
splicing factor proline/glutamine-rich
|
|
chr2_+_96237851
|
0.696
|
|
LOC285033
|
hypothetical protein LOC285033
|
|
chr20_-_31737815
|
0.696
|
NM_005225
|
E2F1
|
E2F transcription factor 1
|
|
chr6_+_24511123
|
0.695
|
NM_020662
|
MRS2
|
MRS2 magnesium homeostasis factor homolog (S. cerevisiae)
|
|
chr1_+_153545162
|
0.693
|
NM_001135822
NM_002004
|
FDPS
|
farnesyl diphosphate synthase
|
|
chr18_-_43710917
|
0.692
|
NM_005901
|
SMAD2
|
SMAD family member 2
|
|
chr16_-_10943778
|
0.691
|
|
DEXI
|
Dexi homolog (mouse)
|
|
chr2_+_223433919
|
0.689
|
NM_004457
NM_203372
|
ACSL3
|
acyl-CoA synthetase long-chain family member 3
|
|
chr19_+_16083549
|
0.683
|
|
RAB8A
|
RAB8A, member RAS oncogene family
|
|
chr1_+_153545301
|
0.677
|
NM_001135821
|
FDPS
|
farnesyl diphosphate synthase
|
|
chr6_-_83014160
|
0.676
|
NM_015525
|
IBTK
|
inhibitor of Bruton agammaglobulinemia tyrosine kinase
|
|
chr2_+_27127994
|
0.676
|
NM_001035507
NM_021831
|
AGBL5
|
ATP/GTP binding protein-like 5
|
|
chrX_+_49013243
|
0.675
|
NM_001184745
NM_033215
|
PPP1R3F
|
protein phosphatase 1, regulatory (inhibitor) subunit 3F
|
|
chr17_-_44642934
|
0.672
|
NM_001198754
|
GNGT2
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2
|
|
chr13_-_48916144
|
0.671
|
NM_001079670
|
CAB39L
|
calcium binding protein 39-like
|
|
chr7_+_133982070
|
0.671
|
NM_001724
NM_199186
|
BPGM
|
2,3-bisphosphoglycerate mutase
|
|
chr2_+_74539039
|
0.670
|
NM_012477
|
WBP1
|
WW domain binding protein 1
|
|
chr6_-_136613097
|
0.670
|
NM_001099286
NM_138419
|
FAM54A
|
family with sequence similarity 54, member A
|
|
chr7_-_137337318
|
0.669
|
|
CREB3L2
|
cAMP responsive element binding protein 3-like 2
|
|
chr1_-_35795592
|
0.666
|
NM_024874
|
KIAA0319L
|
KIAA0319-like
|
|
chr7_+_149166441
|
0.658
|
NM_001099220
|
ZNF862
|
zinc finger protein 862
|
|
chr7_-_72735665
|
0.656
|
NM_032317
|
DNAJC30
|
DnaJ (Hsp40) homolog, subfamily C, member 30
|
|
chr3_-_42820915
|
0.653
|
NM_001099668
|
HIGD1A
|
HIG1 hypoxia inducible domain family, member 1A
|
|
chrX_+_151750166
|
0.652
|
NM_001129765
NM_015922
|
NSDHL
|
NAD(P) dependent steroid dehydrogenase-like
|
|
chr16_-_1816529
|
0.652
|
|
HAGH
|
hydroxyacylglutathione hydrolase
|
|
chr9_-_35805040
|
0.649
|
NM_032593
|
HINT2
|
histidine triad nucleotide binding protein 2
|
|
chr2_+_27128024
|
0.648
|
|
AGBL5
|
ATP/GTP binding protein-like 5
|
|
chr11_-_62355128
|
0.647
|
|
STX5
|
syntaxin 5
|
|
chr14_-_22825072
|
0.647
|
|
HOMEZ
|
homeobox and leucine zipper encoding
|
|
chr1_-_115014151
|
0.647
|
NM_198459
|
DENND2C
|
DENN/MADD domain containing 2C
|
|
chr7_-_137337352
|
0.646
|
|
CREB3L2
|
cAMP responsive element binding protein 3-like 2
|
|
chr1_+_12002138
|
0.643
|
|
MIIP
|
migration and invasion inhibitory protein
|
|
chr11_+_57236617
|
0.643
|
NM_001144012
NM_015959
|
TMX2-CTNND1
TMX2
|
TMX2-CTNND1 read-through transcript
thioredoxin-related transmembrane protein 2
|
|
chr17_+_43373887
|
0.642
|
NM_018129
|
PNPO
|
pyridoxamine 5'-phosphate oxidase
|
|
chr1_-_52116598
|
0.642
|
|
NRD1
|
nardilysin (N-arginine dibasic convertase)
|
|
chr12_+_54904371
|
0.640
|
NM_024068
|
OBFC2B
|
oligonucleotide/oligosaccharide-binding fold containing 2B
|
|
chr10_-_69267774
|
0.640
|
NM_021800
NM_201262
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12
|
|
chr10_-_101370147
|
0.640
|
|
SLC25A28
|
solute carrier family 25, member 28
|
|
chr1_-_226679648
|
0.638
|
NM_003493
|
HIST3H3
|
histone cluster 3, H3
|
|
chr3_+_67131416
|
0.637
|
NM_032505
|
KBTBD8
|
kelch repeat and BTB (POZ) domain containing 8
|
|
chr15_+_39310693
|
0.637
|
NM_007236
|
CHP
|
calcium binding protein P22
|
|
chr12_+_110689094
|
0.636
|
|
ALDH2
|
aldehyde dehydrogenase 2 family (mitochondrial)
|
|
chr1_+_43888382
|
0.634
|
NM_014663
|
KDM4A
|
lysine (K)-specific demethylase 4A
|
|
chr17_+_18700385
|
0.632
|
|
PRPSAP2
|
phosphoribosyl pyrophosphate synthetase-associated protein 2
|
|
chr1_-_23567084
|
0.627
|
NM_030634
|
ZNF436
|
zinc finger protein 436
|
|
chr1_+_165957780
|
0.626
|
NM_001146191
NM_003953
NM_024569
|
MPZL1
|
myelin protein zero-like 1
|
|
chrX_-_46889036
|
0.626
|
NM_001135998
NM_019056
|
NDUFB11
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 11, 17.3kDa
|
|
chr16_+_1817209
|
0.625
|
NM_001018104
NM_001142398
NM_031208
|
FAHD1
|
fumarylacetoacetate hydrolase domain containing 1
|
|
chr20_-_48203709
|
0.622
|
NM_001162505
NM_199129
NM_199203
|
TMEM189
TMEM189-UBE2V1
|
transmembrane protein 189
TMEM189-UBE2V1 readthrough
|
|
chr19_+_2928480
|
0.618
|
NM_001143986
NM_024760
|
TLE6
|
transducin-like enhancer of split 6 (E(sp1) homolog, Drosophila)
|
|
chr1_-_19101603
|
0.618
|
NM_003748
NM_170726
|
ALDH4A1
|
aldehyde dehydrogenase 4 family, member A1
|
|
chr9_+_132990760
|
0.617
|
NM_005085
|
NUP214
|
nucleoporin 214kDa
|
|
chr1_-_35795589
|
0.616
|
|
KIAA0319L
|
KIAA0319-like
|
|
chr9_-_100057313
|
0.616
|
|
TBC1D2
|
TBC1 domain family, member 2
|
|
chr14_+_23710924
|
0.615
|
|
REC8
|
REC8 homolog (yeast)
|
|
chr19_-_17377383
|
0.614
|
NM_004335
|
BST2
|
bone marrow stromal cell antigen 2
|
|
chr17_+_44325127
|
0.614
|
NM_001002027
NM_005175
|
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9)
|
|
chrX_-_100549444
|
0.607
|
|
GLA
|
galactosidase, alpha
|
|
chr1_+_201541269
|
0.606
|
NM_006763
|
BTG2
|
BTG family, member 2
|
|
chr19_-_3922032
|
0.606
|
|
DAPK3
|
death-associated protein kinase 3
|
|
chr11_-_46824364
|
0.605
|
|
CKAP5
|
cytoskeleton associated protein 5
|
|
chr22_-_21193504
|
0.600
|
NM_080764
|
ZNF280B
|
zinc finger protein 280B
|
|
chrX_+_151750221
|
0.599
|
|
NSDHL
|
NAD(P) dependent steroid dehydrogenase-like
|
|
chr2_-_3360467
|
0.598
|
NM_003310
|
TSSC1
|
tumor suppressing subtransferable candidate 1
|
|
chrX_+_38545597
|
0.598
|
NM_001098790
NM_001098791
|
MID1IP1
|
MID1 interacting protein 1 (gastrulation specific G12 homolog (zebrafish))
|
|
chr17_-_35827568
|
0.598
|
NM_001067
|
TOP2A
|
topoisomerase (DNA) II alpha 170kDa
|
|
chr1_-_149586341
|
0.594
|
NM_000449
NM_001025603
|
RFX5
|
regulatory factor X, 5 (influences HLA class II expression)
|
|
chr12_-_118799457
|
0.592
|
NM_007174
|
CIT
|
citron (rho-interacting, serine/threonine kinase 21)
|
|
chr22_-_17815211
|
0.588
|
|
C22orf39
HIRA
|
chromosome 22 open reading frame 39
HIR histone cell cycle regulation defective homolog A (S. cerevisiae)
|
|
chr14_-_34001164
|
0.588
|
NM_138288
|
C14orf147
|
chromosome 14 open reading frame 147
|
|
chr2_-_157890400
|
0.587
|
NM_020711
|
ERMN
|
ermin, ERM-like protein
|
|
chr1_+_166516901
|
0.586
|
NM_005149
|
TBX19
|
T-box 19
|
|
chr22_+_20326572
|
0.585
|
|
SDF2L1
|
stromal cell-derived factor 2-like 1
|
|
chr20_-_5048443
|
0.582
|
NM_182649
|
PCNA
|
proliferating cell nuclear antigen
|
|
chr11_-_46824416
|
0.579
|
NM_001008938
NM_014756
|
CKAP5
|
cytoskeleton associated protein 5
|
|
chr6_-_83014106
|
0.579
|
|
IBTK
|
inhibitor of Bruton agammaglobulinemia tyrosine kinase
|
|
chr14_+_73621408
|
0.578
|
NM_001024674
|
LIN52
|
lin-52 homolog (C. elegans)
|
|
chr19_-_19110268
|
0.576
|
|
TMEM161A
|
transmembrane protein 161A
|
|
chr9_-_35804982
|
0.576
|
|
|
|
|
chr19_+_47416338
|
0.576
|
|
ZNF526
|
zinc finger protein 526
|